Building a lightweight experiment manager
Track your runs with Hamilton and FastUI
In this post, I’ll share how I built a lightweight experimentation manager for Hamilton. This includes a hook to collect metadata and artifacts from each run, and a web app built with FastAPI + FastUI to view and explore results. In this post, you’ll learn about:
Tracking experiments and code versions together
Building a graphical user interface with FastUI
Extending Hamilton with custom plugins
For my master’s thesis, I trained a total of 240 forecast models (2 model architectures * 4 learning tasks * 3 forecast horizons * 10 prediction targets). For each, I tracked metrics, generated visualizations, and conducted post hoc statistical analyses. Now, multiply this by at least 10 to include the iterations over the codebase to optimize models, include new features, and fix bugs. That’s a lot of numbers to keep track of.
What is experimentation?
For data science, machine learning (ML), and large language model (LLM) applications, an iterative process is necessary to improve metrics of interest (e.g., model performance). A run is a single iteration with a set of hyperparameters, input data, and code implementation for which we collect metrics. An experiment is a set of related runs (e.g., same dataset, same model architecture). In addition to metric values, we typically want to store more complex objects such as tables, figures, ML models that we’ll want to inspect for promising runs. The term artifact refers to the set of objects produced during a run.
Note. Here, "experiments" refers to “offline experimentation” where we use already available data to optimize algorithms. In finance, it is often referred to as backtesting. This notion differs from A/B testing, which is “online experimentation”, where different algorithms/processes are deployed and we compare their effects on an outcome.
Why build an experiment manager?
Nowadays, there exist several mature experiment tracking platforms (MLFlow, Weights&Biases, Neptune.ai, CometML, etc.) full of features. For months, I leveraged an experimentation platform that had integrations with the XGBoost and PyTorch Lightning libraries I used for modeling. Yet, there were at least 2 eventual deal breakers:
Linking code version to experiments
Strong coupling with dataflow
1. Linking code version to experiments
Run reproducibility is greatly hindered if metadata and code version are not strongly linked. While experiment tracking tools are excellent at tracking metadata, code versioning is often limited to logging the Git commit SHA, dependencies, and code to launch the run (not any import). Knowing the code version is essential to reproduce results and identify which runs can be compared together.
Now, consider a typical data scientist development process:
There’s a hypothesis you want to test
You implement code for your analysis
You run the analysis
Completes and produces all necessary results
OR Fails at some point and only produces certain artifacts
If it succeeded: return to step 1 and try something new
If it failed: return to step 2 and fix your code
A data scientist would need to make a git commit each time step 3 is reached. This loop can take less than a minute or a week to complete. People will typically commit after succeeding at step 5 with interesting results. The exact code version for all runs in between is unspecified. For my thesis, I would train 3 out of 240 configurations as a sanity check as I made incremental changes, and train the full 240 models when reaching milestones. It’s essential to know the code implementation associated with each run to make meaningful performance comparisons and conclusions.
2. Strong coupling with dataflow
I have used MLFlow and Weights&Biases extensively before. Their “Get started” examples are very simple as they rely on sensible defaults and use “autolog” features. Nonetheless, real projects will require lots of their boilerplate to use. My generic setup included at least: setup_client, setup_experiment, create_parent_run, create_child_run, and collect_metrics. In reality, none of this code helps me test my hypotheses. It’s required to produce good science, but it should be minimized.
Also, by logging artifacts with a framework, you introduce a rampant dependency in your project. All other scripts and notebooks will now need to use framework.xgboost.load_model() or framework.load_table() instead of the more common and flexible xgboost.load_model() and pandas.read_parquet(). I fought with the framework for hours to store a matplotlib Figure and a pandas DataFrame object (which are very common data science artifacts). Some more fiddling was required to load it back into an exploratory notebook or analysis pipeline.
Finding a solution
Given the limitations for linking code and experiments and the coupling with the dataflow of existing tools, I aimed to develop a solution that would:
Precisely version the experiment code and facilitate viewing changes, in addition to tracking metadata (config, inputs, environment, etc.)
Exist in the periphery of my analysis dataflow, so it doesn’t slow down development, and use standard files and formats for artifacts.
Ideally, everything is coded in Python as I am most familiar with its ecosystem and it involves managing a single toolchain.
Ultimately, I believed solving these problems would improve the experiment tracking experience. So I started building!
What is Hamilton?
Before moving forward with the experiment manager, let’s introduce Hamilton, a central component of my stack. I initially adopted Hamilton to facilitate feature engineering with dataframes, and it became central to my 3 analysis pipelines: data preparation, model training, and post hoc statistical testing.
It is a general-purpose framework to write dataflows using regular Python functions. At its core, each function defines a transformation and its parameters indicate its dependencies. Hamilton automatically connects individual functions into a Directed Acyclic Graph (DAG) that can be executed, visualized, optimized, and reported on.
When using Hamilton, you define 1) the individual functions of your dataflow into a Python module and 2) a Driver object that will assemble and execute the dataflow.
My project structure is the following:
masters/
├── scripts/
│ └── train_dl.py
└── src/
├── __init__.py
├── dl_dataflow.py
└── pytorch_models.pyThe complex PyTorch and PyTorch Lightning objects would be defined in pytorch_models.py, the Hamilton dataflow in dl_dataflow.py, and the Driver code to run experiments under the scripts/ directory. Organizing my code this way facilitates the reusability of components, such as loading the dataflow in a notebook or using the PyTorch model outside of Hamilton.
While dl_dataflow.py defines operations, scripts/train_dl.py specifies how it should run. For instance, it provides a clear interface to load the dataflow, pass a configuration and inputs, and specify the values to return from the dataflow.
Here’s an overview of the Driver code:
| from hamilton import driver | |
| from hamilton.io.materialization import to | |
| from hamilton.plugins import h_experiments, pandas_extensions | |
| import analysis # <- your module | |
| tracker_hook = h_experiments.ExperimentTracker( | |
| experiment_name="hello-world", | |
| base_directory="./experiments", | |
| ) | |
| dr = ( | |
| driver.Builder() | |
| .with_modules(analysis) | |
| .with_config(dict(model="linear", preprocess="pca")) | |
| .with_adapters(tracker_hook) # <- the hook | |
| .build() | |
| ) | |
| inputs = dict(n_splits=4) | |
| # materializers to include in run directory | |
| # use a relative path | |
| materializers = [ | |
| to.pickle( | |
| id="trained_model__pickle", | |
| dependencies=["trained_model"], | |
| path="./trained_model.pickle", | |
| ), | |
| to.parquet( | |
| id="X_df__parquet", | |
| dependencies=["X_df"], | |
| path="./X_df.parquet", | |
| ), | |
| to.pickle( | |
| id="out_of_sample_performance__pickle", | |
| dependencies=["out_of_sample_performance"], | |
| path="./out_of_sample_performance.pickle", | |
| ), | |
| ] | |
| # pass explicitly `tracker_hook.run_directory` | |
| # to store the dataflow visualization | |
| dr.visualize_materialization( | |
| *materializers, | |
| inputs=inputs, | |
| output_file_path=f"{tracker_hook.run_directory}/dag.png", | |
| ) | |
| # launch run with .materialize() | |
| dr.materialize(*materializers, inputs=inputs) |
Building the hook
“Experiment tracking”, as the term implies, should exist outside of the dataflow definition and how it is executed, i.e., the experiment. For this purpose, Hamilton provides the Lifecycle hooks abstraction to capture information about the dataflow at execution time (Learn more). Since it relates to how the code runs, the hook is associated with the Driver in scripts/train_dl.py.
To use it, simply import the hook and add it to the Driver definition like this:
from hamilton.plugins import h_experiments
from src import dl_dataflow # load dataflow
hook = h_experiments.ExperimentTracker(...)
dr = (
driver.Builder()
.with_modules(dl_dataflow) # pass dataflow to Driver
.with_adapters(hook) # pass hook to Driver
.build()
)Creating your own hook is actually quite simple. Simply define a new class with methods that you want to have executed at specific points in the graph lifecycle. For instance, the ExperimentTracker hook uses the post graph construction, before/after graph execution, and before/after node execution. The different hooks allow us to version the dataflow code used by the Driver, log the inputs, and store relevant results.
Quickly, another important notion is materialization (Learn more). Essentially, it means loading or storing data in some format. This is great for us because it allows us to specify what experiment artifacts we want to store and how.
Here’s some pseudo-code showcasing the important logic of the hook. Explanations are below, but try reading the code first and see what you understand from it.
| class ExperimentTracker( | |
| lifecycle.NodeExecutionHook, | |
| lifecycle.GraphExecutionHook, | |
| lifecycle.GraphConstructionHook, | |
| ): | |
| def __init__(self, experiment_name: str, base_directory: str): | |
| self.cache = Cache(base_directory) | |
| self.init_directory = Path.cwd() | |
| self.run_directory = resolve_path(base_directory) | |
| self.materialized = list() | |
| def post_graph_construct(self, config): | |
| """Collect the Driver config""" | |
| self.config = config | |
| def run_before_graph_execution(self, run_id, graph, inputs, overrides): | |
| """Gather metadata about the starting run""" | |
| self.run_id = run_id | |
| self.graph_hash = hash(graph) | |
| self.inputs = get_input_nodes(graph.nodes) | |
| self.overrides = overrides | |
| def run_before_node_execution(self, node): | |
| """Move to run directory to produce materialized result""" | |
| if is_materializer(node): | |
| os.chdir(self.run_directory) | |
| def run_after_node_execution(self, node): | |
| """Return to init directory after materialization. Log materialization""" | |
| os.chdir(self.init_directory) | |
| self.materializers.append(node) | |
| def run_after_graph_execution(self): | |
| """Collect metadata stored in tracker and write to cache""" | |
| metadata = get_run_metadata(self) | |
| self.cache.write(self.run_id, metadata) |
Hook logic:
At instantiation, we open our central metadata cache and create the unique run directory for artifacts.
We collect metadata after graph construction (when you do
driver.Builder().build()for those familiar) and before graph execution (afterdr.materialize()). This includes:Driver config (after graph construction)
Unique run id
Hash of the executed code
Inputs
Overrides
For artifacts, it’s a bit tricky to store them in the run directory, which didn’t exist before running the code. To do so, we move to the run directory before a materializer node is executed. After the materializer node is executed, we return to the init directory. This of course is an implementation detail and one of the design choices I made with this approach.
After the graph execution is completed, the run metadata and the paths to artifacts are stored in the central cache.
Benefits
This pattern allowed me to focus on writing my research dataflows and avoid wrestling with the framework (except for building the hook 🤓). If my Hamilton dataflow works, the experiment tracking will work.
The hook creates a directory for each run, preventing me from accidentally overwriting important artifacts (e.g., a model from a previous run).
The dataflow code is hashed/versioned each time the code runs, independently of git commits. It currently hashes the whole graph but could provide diffs at the function level.
Requires Hamilton and no additional dependencies. Using standard file formats for artifacts facilitates interoperability.
Extend it further
I hope this showed the value and how easy it is to define a custom hook. There are many exciting avenues to handle artifacts and capture additional metadata, and even integrate with existing tooling providers in a cleaner way:
Store artifacts in the cloud
Compress artifacts
Execution time
Git SHA
Source code for the dataflow
Source code for the driver and execution
Environment, user, and system information
View the full code for the hook on GitHub to get started!
Building the application
Generating metadata and artifacts is only half the battle. Next, you need a tool to efficiently sift through 100s, if not 1000s, of runs and compare them in a meaningful way. On one hand, you should write code (in a script or notebook) to aggregate, rank, and compare metrics to find the most promising runs. On the other hand, visualizations and tables can better represent multiple dimensions at once and reveal unexpected patterns (especially in failure cases). For this reason, it’s convenient to have a graphical user interface that displays both.
Here are a few key features and design decisions:
Meant as a “read-only” UI of experiments tracked by Hamilton.
Hierarchy between experiment > code version > run
Filter runs by code version
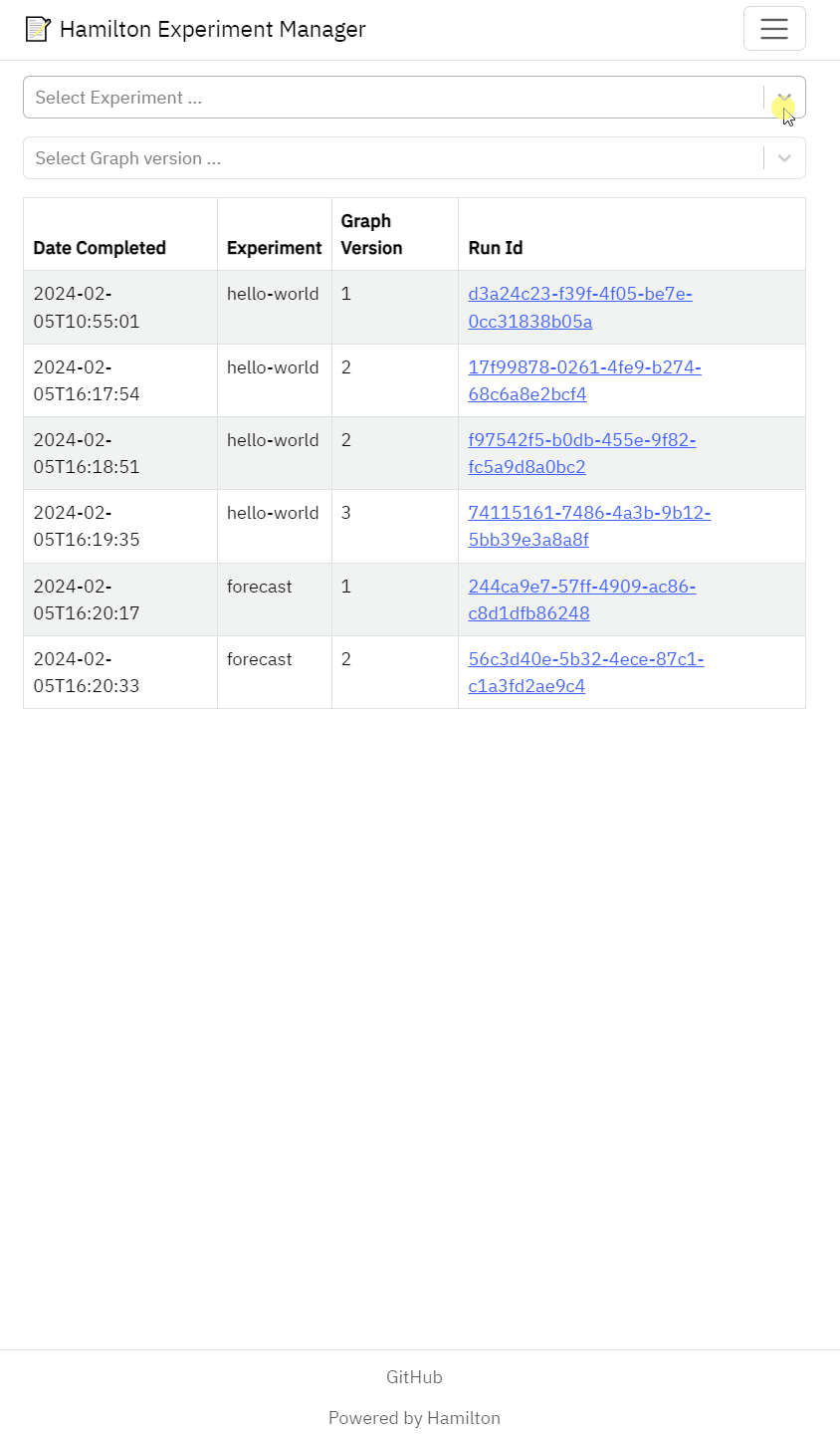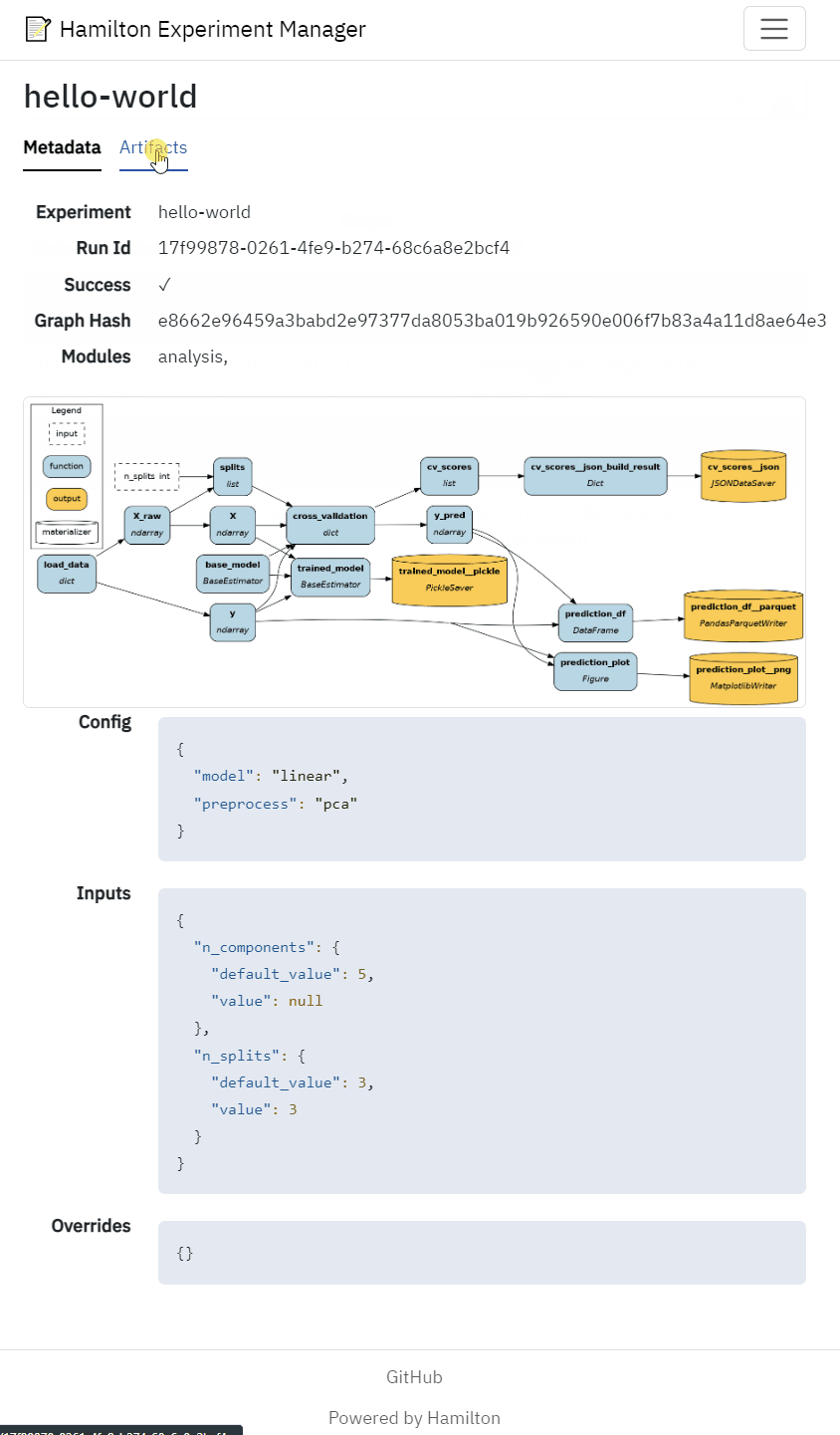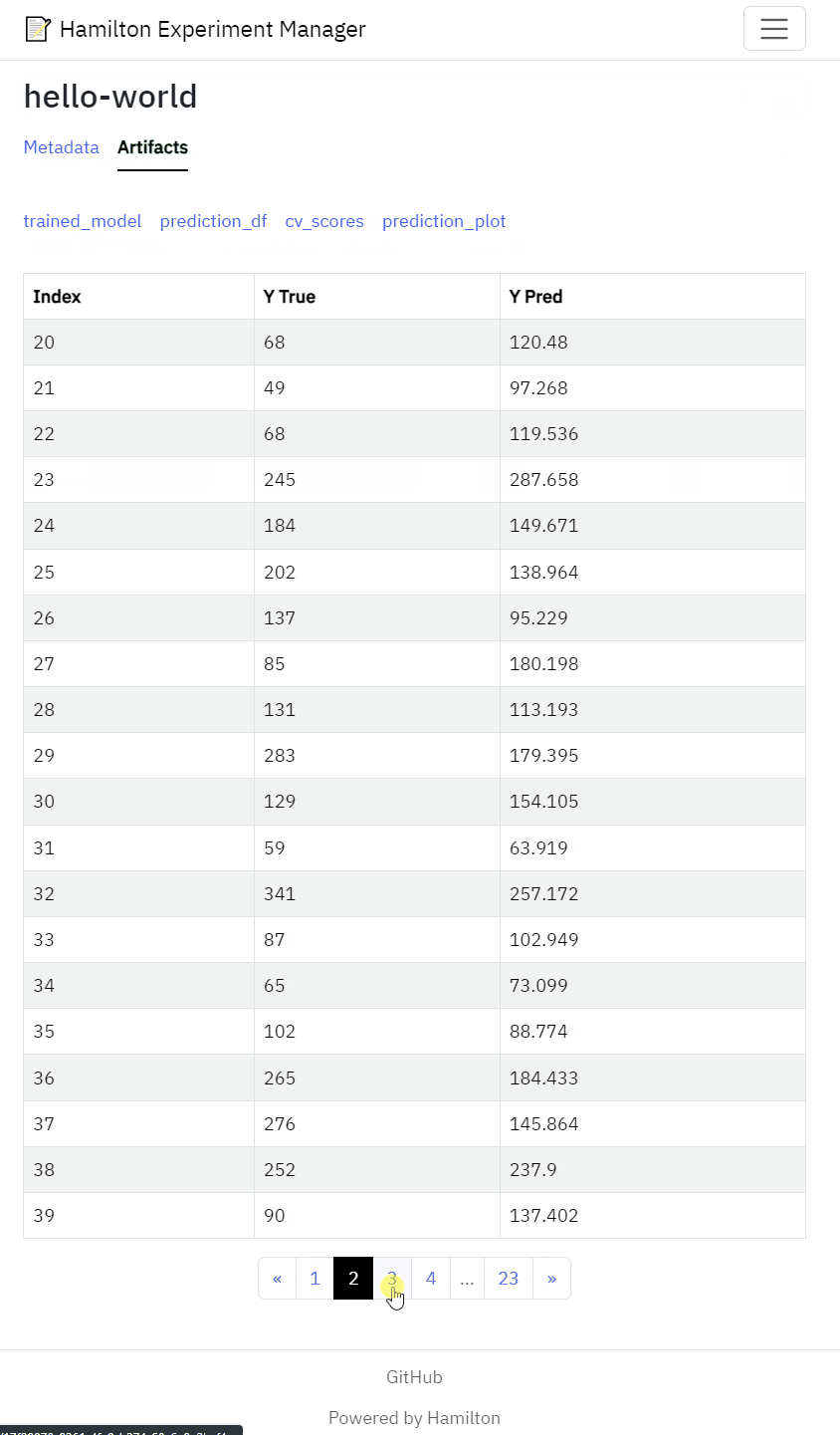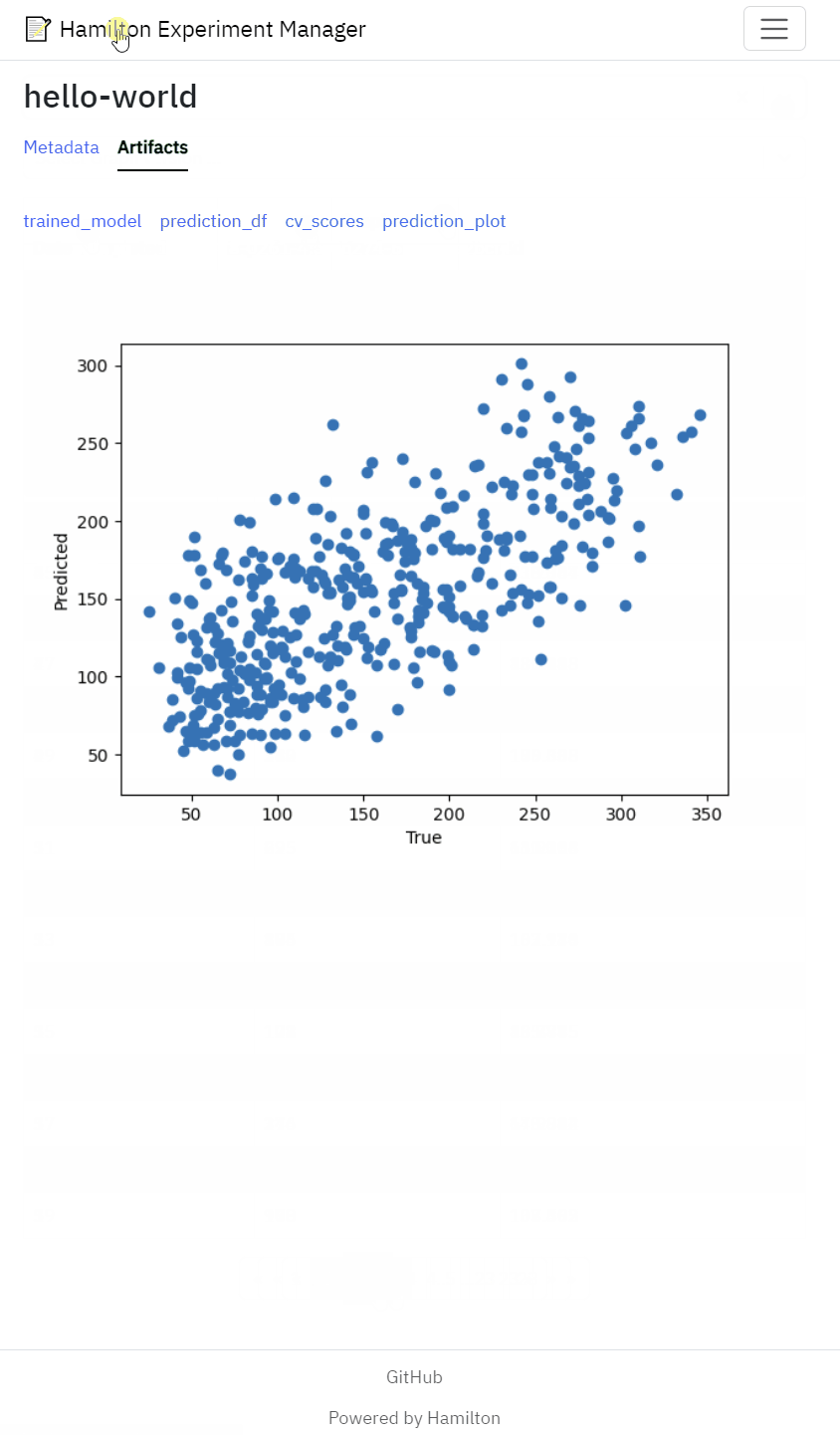
View the dataflow visualization for each Hamilton run
View run metadata (config, inputs, date, etc.)
View artifacts such as performance metrics, statistical test results model performance figures
Lightweight local server with a fast and responsive UI
Streamlit?
A popular option for web applications is Streamlit. It allows you to create a webpage as a Python script (a .py file) that declares and renders UI components from top to bottom. It is remarkable for quickly building proofs-of-concept, but this execution model breaks for complex applications.
First, re-executing the entire page often results in sluggish performance especially when costly I/O operations, data transformations, or API calls are required. Consequently, a lot of explicit state management code needs to be written to improve the user experience. Second, although Streamlit supports multi-page apps, each page is its own .py file (i.e., multiple individual Streamlit pages). This further complicates the necessary state management to communicate between pages and makes code duplication hard to avoid.
I previously built several applications with Streamlit + Hamilton, and it does help with performance, state management, and code reusability. But I always end up wrestling with Streamlit despite my familiarity.
FastUI: a new alternative
FastAPI is a leading Python web framework to define endpoints and create data APIs. Now, the recently released FastUI framework extends FastAPI to enable the creation of a React frontend in Python (intro to React). Essentially, you define a page as a list of UI components, and use API endpoints to interact with the web app (GET to request a page, POST to send user inputs).
See the FastUI demo.
In particular, FastUI’s React-ive execution model provides better performance than Streamlit’s. Also, it benefits from FastAPI’s automatic handling of asynchronous requests. This leads to a very snappy user interface. In addition, building large multi-page applications remains simple since you can rely on FastAPI’s feature to create routes and subroutes. This allows you to organize your codebase across multiple files, in the way that best suits your needs.
Note. FastAPI also has great synergy with Hamilton.
Implementation
To work, the server loads in memory the run metadata cache and mounts the directory storing experiment artifacts.
from fastapi import FastAPI
from fastapi.staticfiles import StaticFiles
app = FastAPI()
app.mount("/experiments", StaticFiles(directory=EXPERIMENT_DIR), name="experiments")
runs = get_runs_from_cache(EXPERIMENT_DIR)Then, endpoints are responsible for manipulating metadata and loading artifacts to create an HTML page to return to the user. Since these HTML pages have the @pydantic/fastui-prebuilt package, interacting with a component (links, buttons, pages, etc.) sends requests to the FastAPI server to update the state of the current page or return a new one. For example, the following code implements the Run metadata page:
| @app.get("/api/run/{run_id}/", response_model=FastUI, response_model_exclude_none=True) | |
| def run_metadata(run_id: str) -> list[AnyComponent]: | |
| run = run_lookup()[run_id] | |
| return base_page( | |
| c.Heading(text=run.experiment, level=2), | |
| *run_tabs(run_id=run_id), | |
| c.Details( | |
| data=run, | |
| fields=[ | |
| DisplayLookup(field="experiment"), | |
| DisplayLookup(field="run_id"), | |
| DisplayLookup(field="success"), | |
| DisplayLookup(field="graph_hash"), | |
| DisplayLookup(field="modules"), | |
| ], | |
| ), | |
| c.Image( | |
| src=f"/experiments/{run.experiment}/{run.run_id}/dag.png", | |
| width="100%", | |
| height="auto", | |
| loading="lazy", | |
| referrer_policy="no-referrer", | |
| class_name="border rounded", | |
| ), | |
| c.Details( | |
| data=run, | |
| fields=[ | |
| DisplayLookup(field="config", mode=DisplayMode.json), | |
| DisplayLookup(field="inputs", mode=DisplayMode.json), | |
| DisplayLookup(field="overrides", mode=DisplayMode.json), | |
| ], | |
| ), | |
| ) |
The FastUI interface is very responsive and feels more robust than Streamlit. Being easy to extend, I can see FastUI applications start as proof-of-concept, and eventually grow into production-ready services and directly integrate into products. Adoption is facilitated if you are familiar with FastAPI and your project already leverages Pydantic. While this project is definitely one to follow, note that the documentation is currently at its slimmest, and there are much fewer UI components available.
Extend it further
FastAPI + FastUI makes it easy to add pages or components to an existing app compared to Streamlit. There are plenty of ways that the experiment server could be improved:
Load cloud artifacts
Display code diffs between runs
Launch runs from the UI (via valid command line arguments)
View the full code for the hook on GitHub to get started!
FastUI is a novel tool with great potential. There’s plenty of room to contribute to it too!
Start tracking your runs!
Adding the Hook
Let’s look at the Driver code to launch a run. The main steps are:
Import the module defining your Hamilton dataflow.
Instantiate the
h_experiments.ExperimentTracker.Build the Driver with the dataflow module and the experiment tracker hook.
Define materializers for artifacts you want to produce. Make sure to use relative paths.
(optional) For visualization functions, which happen outside the execution context, you need to specify the run directory explicitly using
tracker_hook.run_directory.Launch the run with
dr.materialize().
Note that you should capture the visualization before launching the run in case it fails. Also, the way the ExperimentTracker hook is defined, metadata and potentially some artifacts will be captured even if the run fails.
| from hamilton import driver | |
| from hamilton.io.materialization import to | |
| from hamilton.plugins import h_experiments, pandas_extensions | |
| import analysis # <- your module | |
| tracker_hook = h_experiments.ExperimentTracker( | |
| experiment_name="hello-world", | |
| base_directory="./experiments", | |
| ) | |
| dr = ( | |
| driver.Builder() | |
| .with_modules(analysis) | |
| .with_config(dict(model="linear", preprocess="pca")) | |
| .with_adapters(tracker_hook) # <- the hook | |
| .build() | |
| ) | |
| inputs = dict(n_splits=4) | |
| # materializers to include in run directory | |
| # use a relative path | |
| materializers = [ | |
| to.pickle( | |
| id="trained_model__pickle", | |
| dependencies=["trained_model"], | |
| path="./trained_model.pickle", | |
| ), | |
| to.parquet( | |
| id="X_df__parquet", | |
| dependencies=["X_df"], | |
| path="./X_df.parquet", | |
| ), | |
| to.pickle( | |
| id="out_of_sample_performance__pickle", | |
| dependencies=["out_of_sample_performance"], | |
| path="./out_of_sample_performance.pickle", | |
| ), | |
| ] | |
| # pass explicitly `tracker_hook.run_directory` | |
| # to store the dataflow visualization | |
| dr.visualize_materialization( | |
| *materializers, | |
| inputs=inputs, | |
| output_file_path=f"{tracker_hook.run_directory}/dag.png", | |
| ) | |
| # launch run with .materialize() | |
| dr.materialize(*materializers, inputs=inputs) |
Run the server
Once you have the Hamilton package installed with the experiments extras, pip install sf-hamilton[experiments], start the FastAPI server with
h_experimentsYou should see in the terminal:
INFO: Started server process [24113]
INFO: Waiting for application startup.
INFO: Application startup complete.
INFO: Uvicorn running on http://127.0.0.1:8123 (Press CTRL+C to quit)You can specify your experiment directory (default: “./experiments”) and the host and port with:
h_experiments $/PATH/TO/EXPERIMENTS --host $HOST --port $PORTTo see an example in action, checkout this example in the Hamilton repository.
Recap
Code versioning and experiment tracking are essential to any data initiative (DS, ML, LLM). Experiment tracking platforms provide a plethora of features, but there’s still room to improve development ergonomics. Why wait after them? Build it yourself!
In this post, we showed how to create a custom lifecycle hook for Hamilton and developed a web app with FastAPI + FastUI in order to improve experiment versioning, reduce boilerplate code, and improve development ergonomics.
These features are now readily available in Hamilton and yours to try and customize further!
Links
📣 join our community on Slack — we’re more than happy to help answer questions you might have or get you started.
⭐️ us on GitHub
📝 leave us an issue if you find something
Other Hamilton posts you might be interested in:
Learn more about hooks: Customizing Hamilton’s Execution
Learn more about materialization: Separate data I/O from transformation -- your future self will thank you.
Learn more about FastUI: https://github.com/pydantic/FastUI
Datadog hook: Monitoring dataflows with Hamilton + Datadog