Modeling Pregnancy Due Dates with Hamilton
How to build an interactive dataflow to represent conditional probability calculation for an expecting mother's due date
This post was written on July 13th 2024. The baby came on July 22nd. The conditional probability of that was 5.01%, and it was the fourth most-likely date.
You can find the code (notebook + modules) for the blog post here.
The Problem
A baby never comes on time. Well, a baby rarely comes on time. In fact, there is a 3% chance that a baby will come on the due date, a 50.5% chance that it will come before, and a 46.5% chance that it will come after.
For parents expecting to deliver naturally, this can be an unsatisfying answer:
When will my baby be here?
Well, given that today is July 14, your due date is July 18, and the baby has not yet come, here is the skew-normal probability distribution that describes it (big curve). Oh, and if it doesn’t come by the end of July you’ll want to induce.
While I am as frustrated about the lack of certainty as anyone (this lesson applies to more than just simple predictions, by the way), I do have one advantage. I’m an absolute nerd – I have a passing fancy for conditional probabilities and pretty plots, as well as an obsession for building elegant data manipulation systems.
In this post, we’re going to calculate due date probabilities using a fairly simple statistical model. In doing so, we will show you how to use Hamilton for production-ready EDA inside a notebook, utilize the builtin calendar library to visualize our results, and reverse engineer the parameters of a probability distribution in an extremely hacky but simple way. All this because the room is painted, the crib is assembled, the go-bag is packed (well, hers is but I’m procrastinating), and really, there is very little else for me to do.
Hamilton
We will go over visualization/modeling tools when we use them, but let’s explain Hamilton a bit before diving in. Hamilton is a lightweight python library for building dataflows (any computation requiring data) out of python functions. It can run wherever Python does, and is widely used for anything from simple statistical modeling (this post) to more complex ML, to online RAG in web services and more complex ingestion pipelines for AI systems.
The core concept is simple – you write each data transformation step as a single Python function, with the following rules:
The name of the function corresponds to the output variable it computes.
The parameter names (and types) correspond to inputs. These can be either passed-in parameters or names of other upstream functions.
This approach allows you to represent your data in ways that correspond closely to code, are naturally self-documenting, and portable across infrastructure.
After writing your functions (assets), you write a driver that executes them – in most cases this is a simple import/run (specifying the assets you want computed and letting the framework do the rest), but it provides options to customize execution:

Hamilton has a notebook integration that we’ll be using in our post, allowing you to define your modules in a cell and reference them in drivers later. You can do this with the %%cell_to_module command, which will update a variable with a module pointer and plot the dataflow defined by the module.

Once you define your module, you will have access to the module pointer as a variable with the name of the module, allowing you to build the driver, just as we did above. See this post for more information on how the integration works.
Modeling Due Dates
While the underlying probability distribution of delivery is actually very complicated and not well known, according to dataayze (the source for a lot of this post), a skewed-normal distribution is a reasonable representation. The due date is roughly the median, and the left-flank is longer tailed (5th percentile is 4 weeks before the due date, 95th percentile is 2 weeks after). A skewed-normal distribution has three parameters.
The first two are the same a standard normal distribution, although they have different meanings
loc – where the curve is shifted (note due to skewness this is *not* the center of mass)
scale – standard deviation. How wide the curve is.
The final parameter is called “skewness” (a in the code below) – this is how lopsided the curve is. To demonstrate, you can see how the probability density function varies based on the skewness parameter for a simple standard normal distribution centered around 0 with scale of 1.

Once we have the right parameters, we should be able to map the calendar dates to the input and use the probability density function (PDF) to calculate probability of delivery *on* a due-date, as well as the cumulative density function (CDF) to calculate the probability of delivery *before* a due date. Simple, right? Almost – there are two wrinkles:
We don’t have the parameters, and they’re weirdly hard to find (in fact, the papers that everyone cites (duration of human singleton pregnancy and gestational length assignment …) don’t represent it as a skewed distribution and instead attempt to estimate the mean/std dev.
There are two other factors we need to account for
Given that today is July 14th, the probability of delivering prior to today is zero. So, the probabilities for future dates will change (increase – they all have to sum to one, and we cut out a big chunk)
Gestation is generally not considered safe post 42 weeks – current medical practice will almost always induce after that.
While (1) would be solved in a better way by finding the data and doing our own analysis (or, asking the author of that website nicely), we’re going to go with maximum effort! We’ll go over how we solve these problems in the code.
Computing with Hamilton
Determining Model Parameters
Recall we didn’t actually know the parameters to the model? Let’s figure them out! We’re going to use the data here: https://datayze.com/labor-probability-chart and reverse engineer what they did (we also reached out for their parameters to compare our results). This part is classic ugly data parsing, that through Hamilton is well organized into functions – we won’t dig too much into the code, but at a high-level, we will:
Copy/paste the probability data into our notebook in the raw_data function
Compute the raw probabilities, using a lot of funky string parsing (avoiding regexes at all cost)
Resample by generating a reservoir of data with 1000 total items (this is clean as the probabilities in the chart had it down to .1% granularity)
Fit to scipy.stats.skewnorm()
Why the roundabout manner with sampling? Well, really, it’s not the right way to do things. We should be using curve_fit (or some similar capability), or even analytically solve it. That’s hard, but the scipy.stats.skewnorm.fit() function is easy, and it gets us close enough.
The DAG to compute the parameters looks like this – we’ve broken out scale, a, and loc using @extract_fields.

We can run this code as follows:

Which gives us the following parameters:
{
"a": -4.186168447183817,
"scale": 20.670154416450384,
"loc": 294.44465059093034
}
This yields a pretty intuitive set of density functions:This yields a pretty intuitive set of density functions:
Again note that while 40 days is the median, the mode is actually around 41 weeks. This also looks close to the data in the website, meaning we’ve reverse-engineered the distribution correctly!
Managing Dates
Before we get around to computing probabilities using the model above, let’s set up a few date-related variables we can use. In this case, we will model the due date from the start date (the first day of the mother’s period), an input to our dataflow. The due date is defined as 40 weeks past the start_date (a 9-month gestation period + a full cycle).
We restrict our calculation down to just a smaller set of dates (8 weeks before and 4 weeks after), largely to make the data sane. Thus we have possible_dates, which will form the “spine” (common index) for the rest of the data:

This code presents the following (simple) DAG:
Computing Probabilities
Now that we have the base model as well as our dates, let’s actually calculate the probabilities. First we’re going to compute over a larger date range (0->365 days), which we will later truncate. We create a scipy rv_continuous object that allows us to compute the density functions we want. We then do some adjustments to model our initial assumptions – both the current date and the induction date:
This is the meat of the code – we do two adjustments:

We truncate to past the current date (if specified), and readjust the probabilities to sum to 1)
We truncate after the “induction date” (defaults to two weeks post due-date), and add the entire mass to the induction date
While there are obviously a few simplifying assumptions (fixed induction date, quantized date set, etc…), this is a pretty reasonable model.
Next all we need to do is compute the cdf with a cumsum() operation (note we’re not using the rv_continous cdf() capability as we transformed the probability distribution into a discrete function with different values and cumsum() will work just fine. While we could define our own rv_continous variable, the discrete modeling here is a simple, debuggable approach. We truncate our distribution function series to have the right range, and return them as our final variables:

Finally, we can build the driver and use it to return a dataframe – in this case we’re using the PandasDataFrameResult result builder, which handles joining for us.

Voila! We have our final results.
Analyzing the Results
Let’s take a look at what we produced. Using a simple seaborn plot (code in example, left out for now), we can see the following distributions.
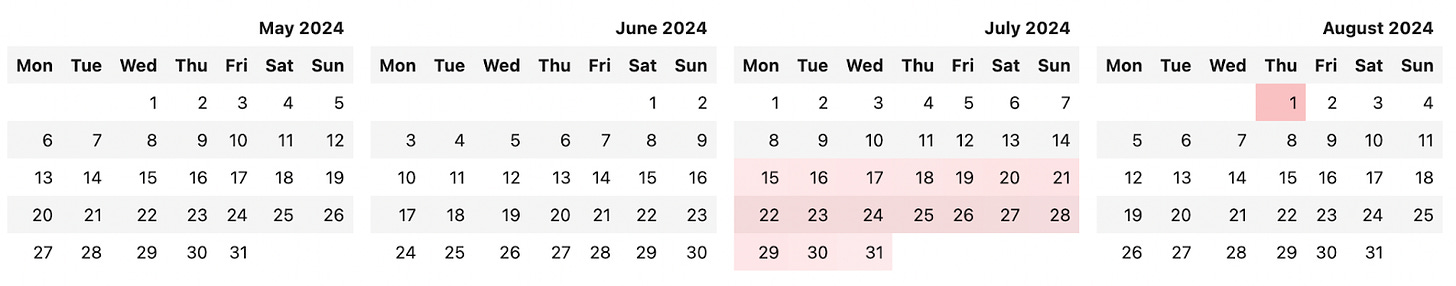
At this point in the pregnancy, it is quite likely that the baby will come late. The mode (obviously) its induction on august 1st, with a 12% chance of that happening. The median is around the 22nd (a week late).
The probability distribution above has no name (of which I’m aware, although the shape is suspiciously close to a common birthing position), but it tells an interesting story. And, it changes every day! Rerun each morning to get your new conditional probabilities.
We can also visualize this in a calendar using the builtin calendar library in python, a tool that enables us to format calendars and perform various calendar-related operations. In this case we’re going to be customizing it heavily – subclassing the HTMLCalendar to allow for highlighting of certain days with a scale:

When we run view_date_range on our data, we get the following:
The real reason I’m doing all of this, is, however, because I want to know the probability that my baby shares a birthday with me! 5% is low, but stranger things than 1/20 have happened.
Notes on Workflow, and Future Reading
All this to determine a 5% probability? Well, the end result doesn’t really tell us much (but hey, it’s a probability distribution, so we’re speaking in possibilities and unknowns anyway). Furthermore, this probably could have been a simpler notebook (avoiding the use of Hamilton all together).
That said, Hamilton made me a lot more productive building this, despite the fact that I was adhering to the rules of a specific framework. I got this productivity gain from the following:
I never worried about notebook state – I had visualizations, and it would recompute whatever needs to be (no changing variable names and accidentally attempting to use them)
I was able to work iteratively – using the Jupyter notebook extensions I can do the following routine
Compute a variable
Play around with it until I’m happy
Tab that cell in (b) in and surround it with a function, put it into the module
Repeat until I’m happy
I get to see the visualization at any point – I can pretty easily figure out what’s happening, what a variable means (that I forgot), etc…
I get code out at the end that’s modular, broken up into functions, and easy to navigate!
This was a toy example. In the real world, there would be a lot more analysis, a lot more spot-checking, and a much more complex set of assumptions to navigate. And that’s where Hamilton really shines – at every stage you’ll have a production-ready ETL to work with. With the Jupyter notebook extension you can even use the -w flag with the %%cell_to_module magic to write your module to a file as you’re editing! This enables you to use the notebook as both a notebook and an IDE.
Learn More
If you’re excited by any of this, or have strong opinions, drop by our Slack channel / leave some comments here! Some resources to help you get started:
📣 join our Hamilton community on Slack — need help with Hamilton? Ask here.
⭐ give hamilton a star on github
📝 open an issue if you find any bugs/want new features
We recently launchedBurr to create LLM agents and applications as well.